¶ Project goals
To bioengineer a strain of Escherichia coli (a.k.a. E. coli) to produce patchoulol, a key ingredient in patchouli fragrance. Other goals are for us Boslab members to learn basic metabolic engineering, document this experience, and produce an instructional module we can share with community DIY and high school biology labs worldwide.
¶ 2020 November 8:
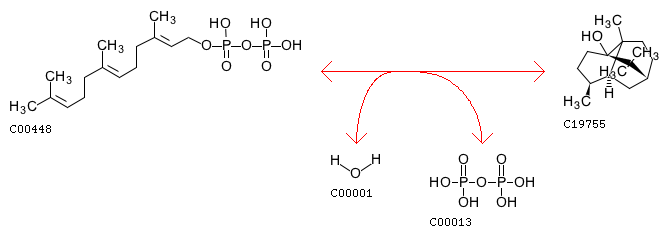
Starting with the KEGG Biochemical Pathway Database, we determined that E. coli has all the elements of a patchoulol production pathway except a gene coding for patchoulol synthase. In particular, E. coli natively produces precursor molecule farnesyl diphosphate (compound C00448 in KEGG). We would only need to add one foreign gene that codes for patchoulol synthase, the enzyme catalyzing the conversion reaction:
Chris Kenyon raised a critical issue: how can we tell whether our gene insertion worked? From the Slack discussion:
What’s our read out? Are we using our noses to detect it? If we have a control strain and smell side by side the patchoulol strain we should be able to detect a difference if we’re reaching threshold. But that’s the question — if we get it work the first try, but at quantities below olfactory threshold, how will we know it worked at all? So try to think about that — what are we going to use as a reporter prior to reaching those necessary levels to actually smell it?
Frank Lee agreed with using our noses for detecting the patchouli smell, and suggested using a colorful protein as a visual reporter for successfully-transformed colony identification on plates (Slack discussion). To ensure that expression of the colorful protein also implies successful expression of patchoulol synthase, he suggested linking the patchoulol synthase coding sequence to the colorful protein coding sequence with a ribosome skip sequence (a.k.a. 2A peptide). He proposed an outline for a possible DNA construct to assemble into a plasmid:
| 5' end | promoter | ribosome binding site | patchoulol synthase coding sequence with missing stop codon | ribosome skip sequence | colorful protein's coding sequence with missing start codon | terminator | 3' end |
How Frank expects this will work:
- The promoter recruits RNA polymerase to transcribe everything between the promoter and terminator into mRNA.
- The mRNA floats around the cytoplasm.
- Through random diffusion, the mRNA encounters a ribosome. The ribosome binds at the ribosome binding site and begins translation, producing the amino acid chain for patchoulol synthase.
- The ribosome encounters the skip sequence, causing it to release the amino acid chain, which folds to become patchoulol synthase enzyme.
- The ribosome continues translation, producing the amino acid chain for the colorful protein.
- Upon encountering the stop codon at the end of the colorful protein's coding sequence, the ribosome releases the amino acid chain, which folds to become the colorful protein that we can see.
Possible problems:
Ribosome skip sequences add a couple amino acids to the end of the first amino acid chain and the beginning of the second chain. This may cause the folded proteins to malfunction.
¶ 2020 November 9:
Chris agreed with this approach (Slack). He suggested using the colorful protein-coding genes from the agar art E. coli strains in the Boslab freezer inventory. Kellen Andrilenas says it's unclear exactly which transgenes are in the agar art strains, but that they're from many years ago when ASM distributed E. coli containing colorful transgenes. (In recent years, ASM has emphasized the use of naturally colorful bacteria, so the 2020 Agar Art competition guide makes no mention of what's in these older strains.) Wendy Pouliot was able to dig up the old ASM Agar Art strain documentation which specify the plasmids they were distributing. Frank tried to summarize:
…the ASM Agar Art plasmids are said to consist of a psb1C2 backbone (typo in the instructions?? psb1C3 exists in Google but not psb1C2) with a BBa_K314100 promoter (iGEM catalog says this is actually a composite part called a "high constitutive expression cassette" consisting of "f1 origin K314110, a high expression promoter J23100, and the Elowitz standard RBS B0034"), and contain one of the following coding sequences:
Pink: BBa_e1010, 706 bp
Blue: BBa_K592009, 669 bp
Teal: BBa_K592011, 702 bp
Orange/Peach: BBa_E2050, 769 bp
Lime Green: BBa_K1033916, 693 bp
Yellow: BBa_E0030, 723 bp
If the backbone is actually pSB1C3, that backbone is 2070 bp.
The BBa_K314100 promoter is 518 bp.
Kellen suggests restriction mapping to verify the constructs in the Agar Art strains we have at Boslab.
¶ 2020 December 13:
In reading about ribosome skip sequences, Frank noticed that they are said to only work in eukaryotes, not prokaryotes. So they won't work in E. coli. Further Googling found the following ResearchGate forum post which suggests that prokaryotic ribosomes can translate multiple open reading frames on a single mRNA, leading to the following revised outline for a DNA construct to assemble for our project:
| 5' end | promoter | ribosome binding site | patchoulol synthase coding sequence | ribosome binding site | colorful protein's coding sequence | terminator | 3' end |
¶ 2021 June 22
At yesterday's Boslab journal club, Chris mentioned that we can order free plasmids from Freegenes.org encoding standard biotech tools such as colorful or fluorescent proteins.
¶ 2021 September 18
Project planning discussion with Chris Kenyon and Ross Mattingly at BosLab. Rough sketch of plan:
- Total RNA extraction
- Freeze patchouli leaf in liquid nitrogen. Pulverize in mortar and pestle.
- Resuspend in TE buffer / H2O.
- Load RNA binding column, elute RNA.
- cDNA synthesis
- Use Chris' kit. And/or try NEB cDNA library kit.
- PCR amplify only the patchoulol synthase cDNA using gene-specific primers.
- Clone into vector of interest.
- Transform vector into E. coli.